Example 2. Visualitzation of globals analysis based in xCGy fragments
The example shows the procedure to extract information from the Global Analyses section of BIGNASim portal. The simple study shown here can be extended to a real use case: the importance of flanking nucleotides in the flexibility of base-pair steps. The effect of the tetranucleotide environment in the sequence-dependent polymorphism of particular base-pair steps has been the target of recent studies. The CG base pair step used for example, shows an interesting bimodal behaviour in one of the six helical base-pair step parameters: Twist (11). In the study, authors claim that the effect of the flanking bases in the CG base pair step is crucial for the existence of two different conformers: High Twist (HT: ~40º) and Low Twist (LT: ~ 20º). Behaviors for each of the 16 possible tetramers including CG are reported. To illustrate the power of BIGNASim database and its interface, two analysis have been chosen: ACGC showing almost no bimodality, and GCGA showing a clear bimodality. The first step uses the Search section of the portal → Search by sequence (GCGA).
Javascript equivalent code
// Direct search of GCGA fragments on both strands
SimulationList = db.simData.find({$or: [{'sequence':/GCGA/}, {'rev-sequence': /GCGA/}]})
// Alternatively search both on only one strand using complementarity
SimulationList = db.simData.find({'sequence':/(GCGA|TCGC)/})
In this case, more than 40 simulations containing this particular fragment are available for selection.
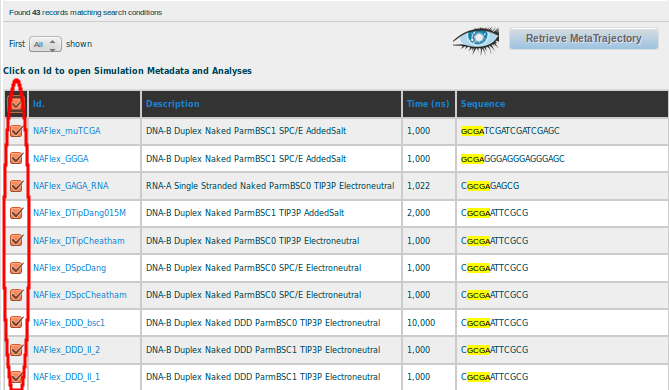
Retrieve Analysis for the selected simulations at the bottom of the page, leads to a Global Analyses page, showing the results for the particular GCGA fragment. Since the interest is studying the possible bimodality showed by the CG base pair step in its Twist parameter when it is surrounded by G and A (GCGA), the CG button should be selected:

Javascript code hint
// Available data for all CpG bpstep (in any simulation, and any sequence
// position) can be retrieved at using just its idGroup: CGCG
DataforAllCpG = db.analData.find( {'_id.idGroup' : 'CGCG} )
// For a simulation SIM and position POS
Datafor1CpG = db.analData.find( {'_id.idSim': SIM, '_id.nGroup': POS, '_id.idGroup' : 'CGCG} )
Twist data can be obtained from "Curves → Helical_bpstep":


BIGNASim in its current version contains two kinds of analysis for each of the six helical base-pair step parameters: one with the values for every snapshot of all the selected simulations, and one with the time-averaged values for each simulation. To show the bimodality, histogram with all the values for the Twist parameter should be chosen:

In the histogram plot, the average is represented as a vertical blue line, and the experimental value, used as reference, is represented as a vertical red line (see Example of use 5 for a detailed description of the use of experimental data).

Javascript code
// Code to retrieve twist values for a complete trajectory for a given Simulation SIM, CpG POS
twistData_C = db.analData.find(
{'_id.idSim': SIM, '_id.nGroup': POS, '_id.idGroup': 'CGCG'}
).sort('_id.frame':1};
while (twistData_C.hasNext() {
Data = twistData_c.next();
printjson (Data._id.frame + ' ' + Data.CURVES.helical_bpstep.twist);
}
The histogram shows two well defined populations, centered at ~25º and ~35º, in good agreement with the previously presented study (11). To analyse the influence of the surrounding bases in the CG base pair step (tetramer influence), the procedure will be repeated seeking for the fragment ACGC. (Search by Sequence → Select All → Open Analysis for the selected simulations).

And CG → Curves → Helical_bpstep Analysis → Twist Analysis.

The new histogram do seems to follow a normal distribution, although a small shoulder to the low twist conformation can still be identified. The clear difference between the two plots shows that the GCGA tetramer shows a clear bimodality, whereas the ACGC tetramer is more inclined to be in a High Twist conformation. Additionally, raw histogram data can be downloaded for further analysis.
Complete Javascript code to retrieve twist data from ACGC tetramers
SEQ = 'ACGC';
RSEQ = 'GCGT';
// Search for simulations bearing ACGC
SimualtionList = db.simData.find(
{$or: [
{'sequence': {$regex: SEQ}},
{'rev-sequence': {$regex: RSEQ}},
{'_id':1}
]}
).toArray();
// search for CpG fragments
FragsList = db.groupDef.find(
{'_id.idSim': {$in: SimulationList}, 'class': 'CGCG'}
).toArray()
// Iterate over fragments
for (i=0; i < FragsList.length; i++) {
twistData_c = db.analData.find(
{'_id.idSim': FragsList[i]._id.idSim,
'_id.nGroup': FragsList[i]._id.n,
'_id.idGroup' : 'CGCG}
).sort('_id.frame':1};
while (twistData_C.hasNext() {
Data = twistData_c.next();
printjson (Data._id.frame + ' ' + Data.CURVES.helical_bpstep.twist);
}
}